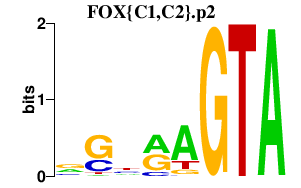
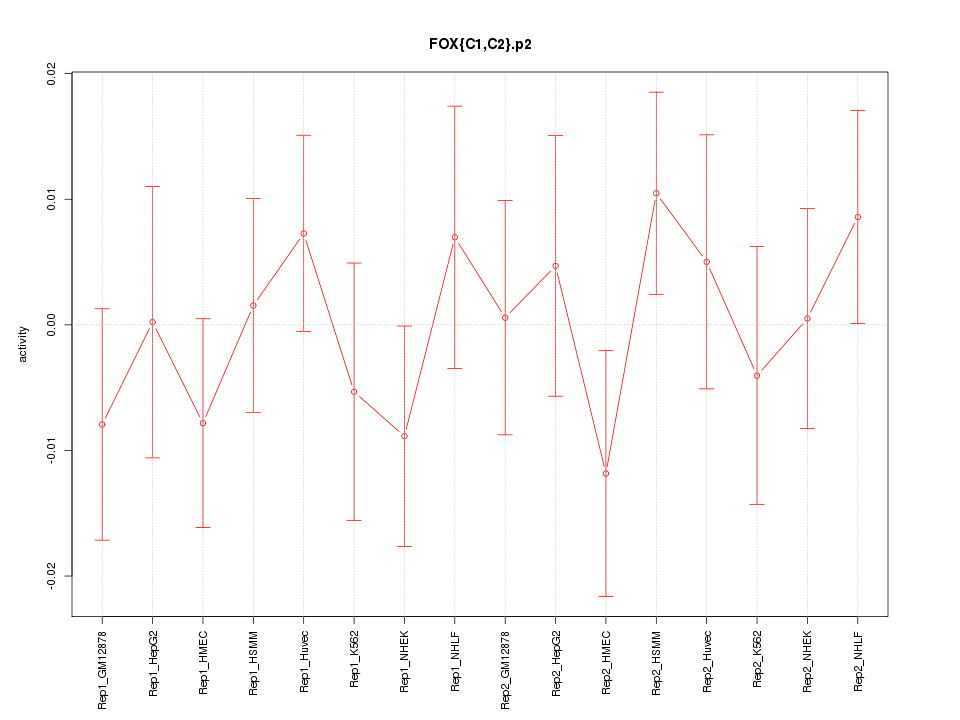
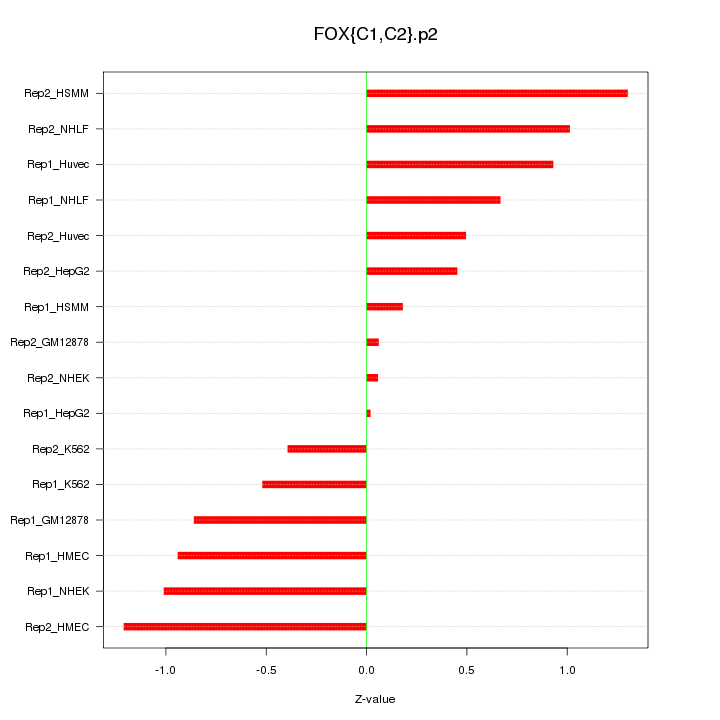
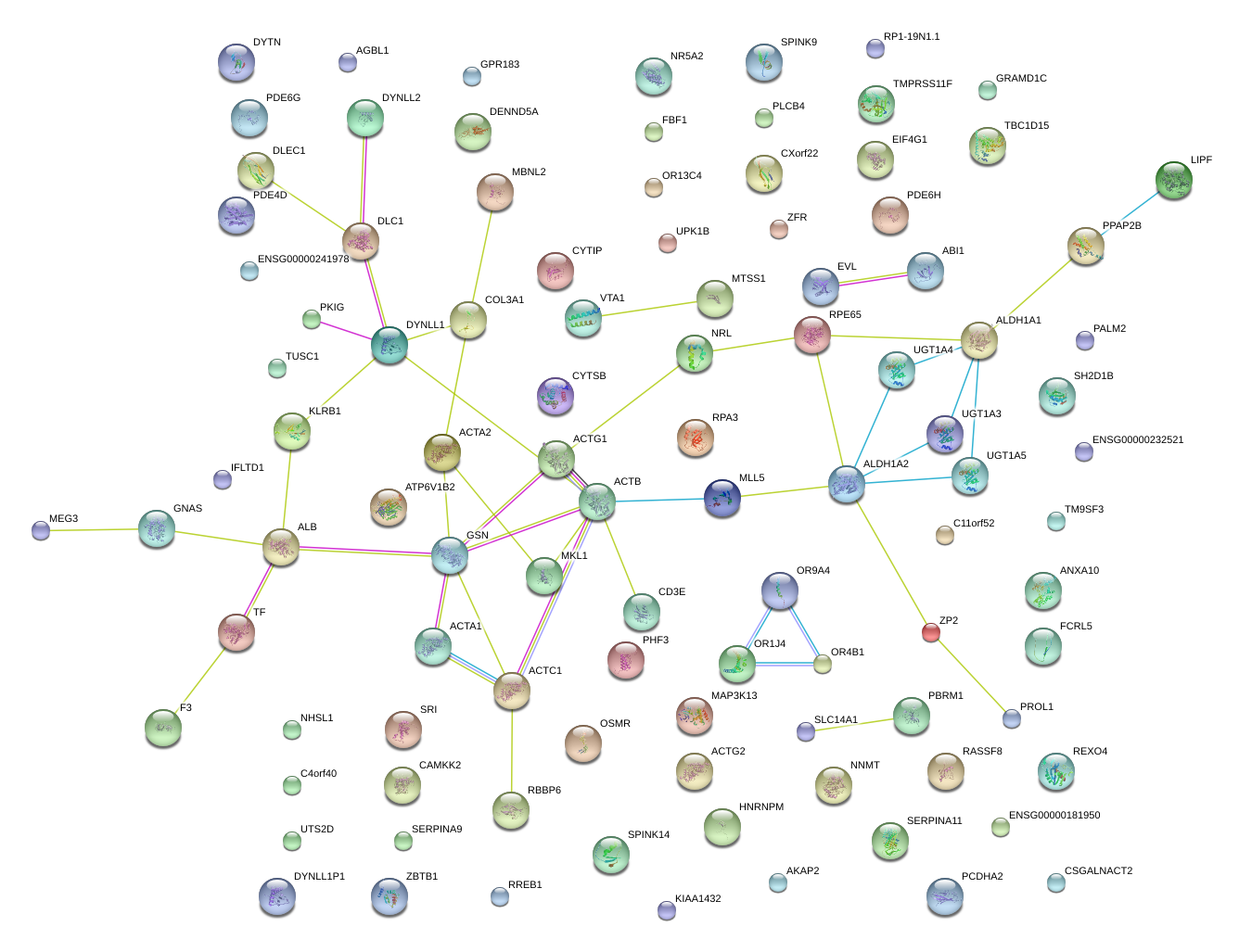
|
chr3_+_120375053
|
0.854
|
NM_006952
|
UPK1B
|
uroplakin 1B
|
|
chr17_-_45626364
|
0.817
|
|
|
|
|
chr8_-_125785611
|
0.815
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr10_-_27097782
|
0.758
|
|
ABI1
|
abl-interactor 1
|
|
chr20_+_56904042
|
0.735
|
|
GNAS
|
GNAS complex locus
|
|
chr9_-_74735669
|
0.730
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr8_+_20099179
|
0.708
|
|
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2
|
|
chr8_+_20099169
|
0.663
|
|
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2
|
|
chr10_-_98315090
|
0.650
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr14_+_64041257
|
0.630
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr2_-_158008763
|
0.619
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr20_+_9146036
|
0.573
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr15_+_84486245
|
0.567
|
NM_152336
|
AGBL1
|
ATP/GTP binding protein-like 1
|
|
chr1_-_56750235
|
0.520
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_-_68688219
|
0.505
|
NM_000329
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa
|
|
chr15_-_56093477
|
0.494
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr14_+_99601498
|
0.470
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr3_-_52688768
|
0.465
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr14_+_99601517
|
0.449
|
|
EVL
|
Enah/Vasp-like
|
|
chr11_+_113672407
|
0.447
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr5_-_58918031
|
0.434
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_147695314
|
0.428
|
NM_001040433
|
SPINK9
|
serine peptidase inhibitor, Kazal type 9
|
|
chr11_-_9182356
|
0.427
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr7_-_87694218
|
0.421
|
NM_198901
|
SRI
|
sorcin
|
|
chr6_-_138862271
|
0.416
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr9_+_5646582
|
0.412
|
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr2_+_73973600
|
0.404
|
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr8_-_13018123
|
0.401
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr5_+_147529488
|
0.400
|
NM_001001325
|
SPINK14
|
serine peptidase inhibitor, Kazal type 14 (putative)
|
|
chr17_-_45620313
|
0.376
|
|
|
|
|
chr13_-_98757634
|
0.371
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr4_+_74500748
|
0.370
|
|
ALB
|
albumin
|
|
chr20_+_42644633
|
0.366
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr2_+_73973640
|
0.361
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr4_+_71298187
|
0.349
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr7_-_7724762
|
0.346
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr11_+_113672378
|
0.331
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr17_-_45626967
|
0.321
|
|
|
|
|
chr5_-_32439067
|
0.317
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr3_-_192531018
|
0.290
|
NM_198152
|
UTS2D
|
urotensin 2 domain containing
|
|
chr20_+_56903595
|
0.282
|
|
GNAS
|
GNAS complex locus
|
|
chr9_+_123088217
|
0.281
|
|
GSN
|
gelsolin
|
|
chr3_+_134978615
|
0.280
|
|
TF
|
transferrin
|
|
chr7_+_141265085
|
0.276
|
NM_001001656
|
OR9A4
|
olfactory receptor, family 9, subfamily A, member 4
|
|
chr22_-_39189369
|
0.274
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr10_+_42953886
|
0.273
|
NM_018590
|
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2
|
|
chr3_+_185516630
|
0.272
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_25597444
|
0.259
|
NM_152590
NM_001145728
NM_001145729
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr6_+_7053183
|
0.256
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr6_-_138862273
|
0.253
|
|
|
|
|
chr6_+_142510108
|
0.246
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr18_+_41558141
|
0.243
|
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr14_-_23623657
|
0.242
|
NM_006177
|
NRL
|
neural retina leucine zipper
|
|
chr3_+_186529376
|
0.238
|
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr16_+_24475129
|
0.235
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr11_+_65027721
|
0.233
|
|
|
|
|
chr12_+_15017222
|
0.231
|
NM_006205
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma
|
|
chr4_-_68678181
|
0.229
|
NM_207407
|
TMPRSS11F
|
transmembrane protease, serine 11F
|
|
chr14_-_93988838
|
0.227
|
NM_001080451
|
SERPINA11
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11
|
|
chr4_+_71054492
|
0.223
|
NM_214711
|
C4orf40
|
chromosome 4 open reading frame 40
|
|
chr9_+_124321239
|
0.219
|
NM_001004452
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4
|
|
chr17_+_19917420
|
0.217
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr6_+_142510075
|
0.217
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr5_+_38881719
|
0.216
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr6_+_142510094
|
0.214
|
NM_016485
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr3_+_115099007
|
0.213
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr6_+_142510034
|
0.211
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr2_+_189547323
|
0.211
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_198263352
|
0.208
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr7_+_104468614
|
0.207
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr11_+_48194937
|
0.207
|
NM_001005470
|
OR4B1
|
olfactory receptor, family 4, subfamily B, member 1
|
|
chr2_+_189547358
|
0.207
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr4_+_169250281
|
0.206
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr9_-_25668855
|
0.201
|
NM_001004125
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr2_-_207291364
|
0.199
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr14_+_64041170
|
0.198
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr14_+_100365162
|
0.197
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr14_+_22059212
|
0.194
|
|
|
|
|
chr2_+_87536174
|
0.194
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr2_+_234286376
|
0.193
|
NM_019078
|
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr10_+_90414125
|
0.193
|
NM_001198828
NM_001198829
NM_001198830
NM_004190
|
LIPF
|
lipase, gastric
|
|
chr1_-_155788775
|
0.191
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr13_+_96672705
|
0.190
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_-_160648429
|
0.185
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr9_+_5619326
|
0.185
|
NM_001135920
|
KIAA1432
|
KIAA1432
|
|
chr16_+_54158084
|
0.183
|
NM_032330
|
CAPNS2
|
calpain, small subunit 2
|
|
chr9_-_106329310
|
0.180
|
NM_001001919
|
OR13C4
|
olfactory receptor, family 13, subfamily C, member 4
|
|
chr11_+_111294868
|
0.178
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr19_+_8415875
|
0.177
|
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr2_+_87536106
|
0.176
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr12_-_120196363
|
0.176
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr12_+_26003218
|
0.175
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr12_+_70519753
|
0.174
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr10_-_112668892
|
0.173
|
NM_001195304
NM_001195305
NM_001195307
|
BBIP1
|
BBSome interacting protein 1
|
|
chr4_+_74491272
|
0.173
|
|
ALB
|
albumin
|
|
chr6_+_64414389
|
0.173
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr16_-_21130368
|
0.171
|
NM_003460
|
ZP2
|
zona pellucida glycoprotein 2 (sperm receptor)
|
|
chr19_+_8415869
|
0.171
|
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr8_+_20098983
|
0.170
|
NM_001693
|
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2
|
|
chr5_+_140154627
|
0.170
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr9_+_111442888
|
0.169
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr11_+_117680504
|
0.168
|
NM_000733
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex)
|
|
chr19_+_8415859
|
0.166
|
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr12_+_25096755
|
0.166
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr5_+_150000394
|
0.166
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr1_+_26358097
|
0.165
|
NM_024869
|
GRRP1
|
glycine/arginine rich protein 1
|
|
chr19_+_53520632
|
0.164
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr8_+_104900591
|
0.161
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr18_-_55177430
|
0.161
|
NM_005570
|
LMAN1
|
lectin, mannose-binding, 1
|
|
chr15_+_55671393
|
0.161
|
NM_001018090
NM_001018091
NM_001018100
NM_152451
|
GCOM1
|
GRINL1A complex locus
|
|
chr16_+_68238460
|
0.158
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr13_-_19244398
|
0.157
|
|
PSPC1
|
paraspeckle component 1
|
|
chr11_+_110890719
|
0.157
|
NM_001100388
NM_207430
|
C11orf88
|
chromosome 11 open reading frame 88
|
|
chr2_+_189547377
|
0.157
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_+_25096472
|
0.156
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_-_54522881
|
0.155
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr1_+_94656517
|
0.155
|
NM_001122674
NM_002858
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3
|
|
chr3_-_158700036
|
0.154
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr9_+_124526089
|
0.154
|
NM_001005235
|
OR1L4
|
olfactory receptor, family 1, subfamily L, member 4
|
|
chr11_-_61823078
|
0.153
|
NM_206998
|
SCGB1D4
|
secretoglobin, family 1D, member 4
|
|
chr11_-_8789457
|
0.153
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr19_+_8415802
|
0.151
|
NM_005968
NM_031203
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr9_-_34719463
|
0.150
|
NM_001141917
|
C9orf144B
|
transmembrane protein C9orf144B
|
|
chr22_-_35970230
|
0.149
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr21_-_37366684
|
0.148
|
NM_153681
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P
|
|
chr20_-_1321571
|
0.145
|
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr14_+_23527866
|
0.142
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chr18_-_63333487
|
0.142
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr3_+_195336625
|
0.139
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr11_-_13474101
|
0.137
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr19_-_47723183
|
0.137
|
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr11_-_123182266
|
0.133
|
NM_001005325
|
OR6M1
|
olfactory receptor, family 6, subfamily M, member 1
|
|
chr15_-_41381744
|
0.132
|
NM_052955
|
TGM7
|
transglutaminase 7
|
|
chr2_+_137464931
|
0.132
|
NM_001080427
|
THSD7B
|
thrombospondin, type I, domain containing 7B
|
|
chr3_-_116272911
|
0.131
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr22_-_35970133
|
0.131
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr6_+_88026338
|
0.131
|
|
ZNF292
|
zinc finger protein 292
|
|
chr12_-_47735373
|
0.130
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr2_-_215998181
|
0.130
|
|
FN1
|
fibronectin 1
|
|
chr11_+_55351270
|
0.129
|
NM_001004739
|
OR5L2
|
olfactory receptor, family 5, subfamily L, member 2
|
|
chr7_+_99263571
|
0.127
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr9_-_130683997
|
0.127
|
NM_001122671
NM_001122672
NM_004059
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic
|
|
chr5_-_156525756
|
0.127
|
NM_130899
|
FAM71B
|
family with sequence similarity 71, member B
|
|
chr2_+_48649662
|
0.126
|
NM_172311
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr6_+_72982865
|
0.126
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr12_+_45896511
|
0.126
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr2_-_233349469
|
0.125
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr5_+_96064248
|
0.125
|
NM_173060
|
CAST
|
calpastatin
|
|
chr11_-_2113297
|
0.124
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr14_+_73884947
|
0.124
|
|
VRTN
|
vertebrae development homolog (pig)
|
|
chr8_+_103633023
|
0.124
|
NM_024410
|
ODF1
|
outer dense fiber of sperm tails 1
|
|
chr3_-_120761092
|
0.123
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr19_+_15713200
|
0.123
|
NM_013938
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3
|
|
chr11_+_111294810
|
0.122
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr16_+_85158357
|
0.121
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr19_-_9087438
|
0.121
|
NM_001005192
|
OR7G1
|
olfactory receptor, family 7, subfamily G, member 1
|
|
chr22_+_38652555
|
0.120
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr9_-_112139908
|
0.120
|
NM_001003936
|
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa)
|
|
chr20_-_14266200
|
0.120
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr2_-_88066423
|
0.119
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr16_-_52295214
|
0.119
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr12_+_115455579
|
0.119
|
|
NCRNA00173
|
non-protein coding RNA 173
|
|
chr5_-_88004891
|
0.118
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr14_+_23849196
|
0.117
|
NM_001164692
NM_019839
|
LTB4R2
|
leukotriene B4 receptor 2
|
|
chr2_+_69055208
|
0.117
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr2_-_74424041
|
0.116
|
NM_133478
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chr5_+_156752739
|
0.115
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr18_-_53438854
|
0.115
|
|
|
|
|
chr14_+_22059174
|
0.113
|
|
|
|
|
chr16_+_68157611
|
0.113
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr10_-_91393610
|
0.113
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr14_+_50096603
|
0.113
|
|
ATL1
|
atlastin GTPase 1
|
|
chr9_+_130684189
|
0.113
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr5_+_43645018
|
0.109
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr12_+_58276114
|
0.109
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chrX_+_15435351
|
0.107
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr1_-_154665807
|
0.107
|
NM_006365
|
C1orf61
|
chromosome 1 open reading frame 61
|
|
chr2_+_132196533
|
0.107
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr9_+_133295297
|
0.106
|
NM_013318
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chr7_+_94123672
|
0.106
|
|
PEG10
|
paternally expressed 10
|
|
chr12_-_55316350
|
0.105
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr1_-_156923111
|
0.105
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr10_+_111755715
|
0.105
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr2_+_158666627
|
0.104
|
NM_173355
|
UPP2
|
uridine phosphorylase 2
|
|
chr6_+_83834080
|
0.104
|
NM_015018
|
DOPEY1
|
dopey family member 1
|
|
chr5_+_112071087
|
0.103
|
NM_001127511
|
APC
|
adenomatous polyposis coli
|
|
chr2_-_224612177
|
0.103
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr5_+_159368690
|
0.102
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr11_+_120478584
|
0.102
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr3_+_195336631
|
0.102
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr4_-_90197140
|
0.102
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr9_-_15240113
|
0.102
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr20_-_1321744
|
0.101
|
NM_000801
NM_054014
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr17_-_64649608
|
0.101
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr1_+_62675296
|
0.101
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr12_+_54231249
|
0.100
|
NM_001005494
|
OR6C4
|
olfactory receptor, family 6, subfamily C, member 4
|
|
chr17_+_70964258
|
0.100
|
NM_014738
|
KIAA0195
|
KIAA0195
|
|
chr15_+_97010219
|
0.099
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr1_+_151081953
|
0.099
|
NM_001128600
|
LCE6A
|
late cornified envelope 6A
|
|
chr14_+_73884918
|
0.098
|
NM_018228
|
VRTN
|
vertebrae development homolog (pig)
|